BioXM™
Knowledge Management Environment
The BioXM™ Knowledge Management Environment is a fully customizable, all-in-one enterprise knowledge management system for better integrated, evidence-based research and development.
Take a look
Take a look at BioXM™
Learn more
The BioXM platform provides a central inventory of all the information representing an organization’s collective knowledge. While individuals focus on generating knowledge in their specific fields of expertise, the whole organization can benefit from that knowledge to make better decisions and follow more intelligent strategies.
The BioXM platform allows quick configuration of highly customized solutions that are driven by specific research and business needs. The knowledge model can continuously adapt to new information or circumstances, which leads to more agility in the face of change, a higher ROI, and ultimately more stability and success.
Your benefits
Your Benefits
The possibilities are virtually limitless. The following are only a few examples.
- Connect and explore clinical data, study information and statistical analyses
- Keep non-redundant repositories of scientific data such as: experimental results, a corporate gene index, a patient register, a phenotype catalog, or a compound and drug database
- Create comprehensive disease maps based on automatic literature mining results
- Connect, organize, review, compare, annotate and modify biological pathways originating from different sources
- Identify biomarker candidates based on automatic literature analysis and compare the results to experimental findings in one platform
- Classify biological functions using ontologies specific to focused scientific areas
- Easily integrate external statistical packages (R, BioConductor), bioinformatics tools, chemoinformatics algorithms, Web services and more
- Generate networks from multiple sources and find previously undetected links in the data
- Keep track of ongoing projects with one click to generate smart folders, table reports and overview graphs
More details
Consolidate knowledge
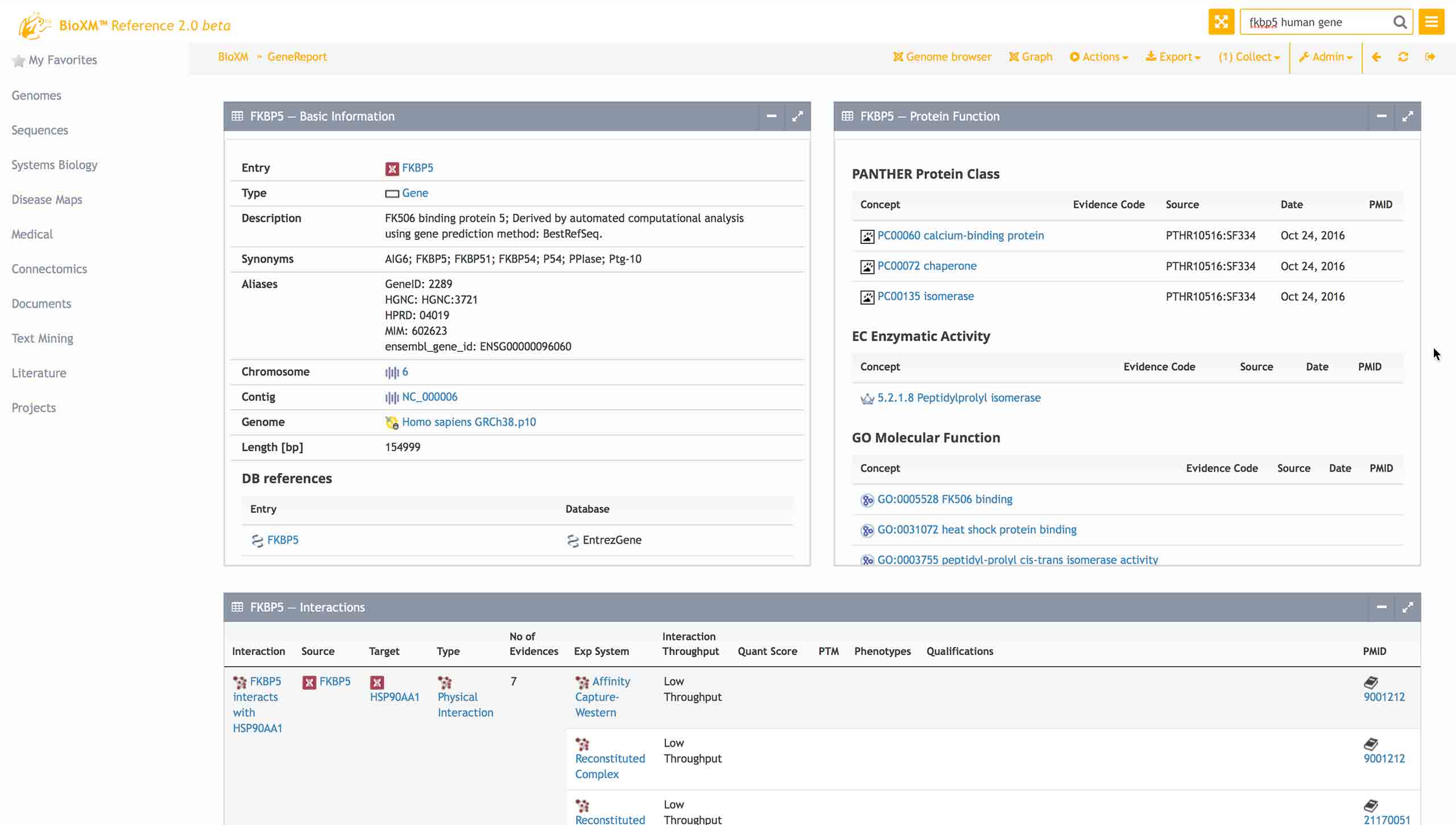
The BioXM system is a project-centered, distributed platform that allows isolated sources of information to be connected into a central inventory of information and knowledge.
Users create, manage and visualize scientific models as extendible networks representing an area of research or a particular perspective.
Reveal hidden relationships
The BioXM platform is configured to the way real users work and real projects develop. It connects the dots between the data and the people behind the data.
Through semantic modeling of interrelated concepts, all perspectives are integrated into the knowledge network and relationships that were previously hidden become clear.
Leverage many perspectives
Personalized user interfaces and flexible access support diverse users and project groups.
Individuals can focus on their science and generating knowledge in their field of expertise while the whole organization can deploy it to gain insight, make better decisions and innovate faster.
Ensure future agility
The BioXM platform can be used to build highly custom solutions through configuration rather than classical software development.
This novel process enables faster, less expensive development driven by real research and business needs.
A solution based on the BioXM platform can, by design, continuously adapt to changing requirements.
Empower end users
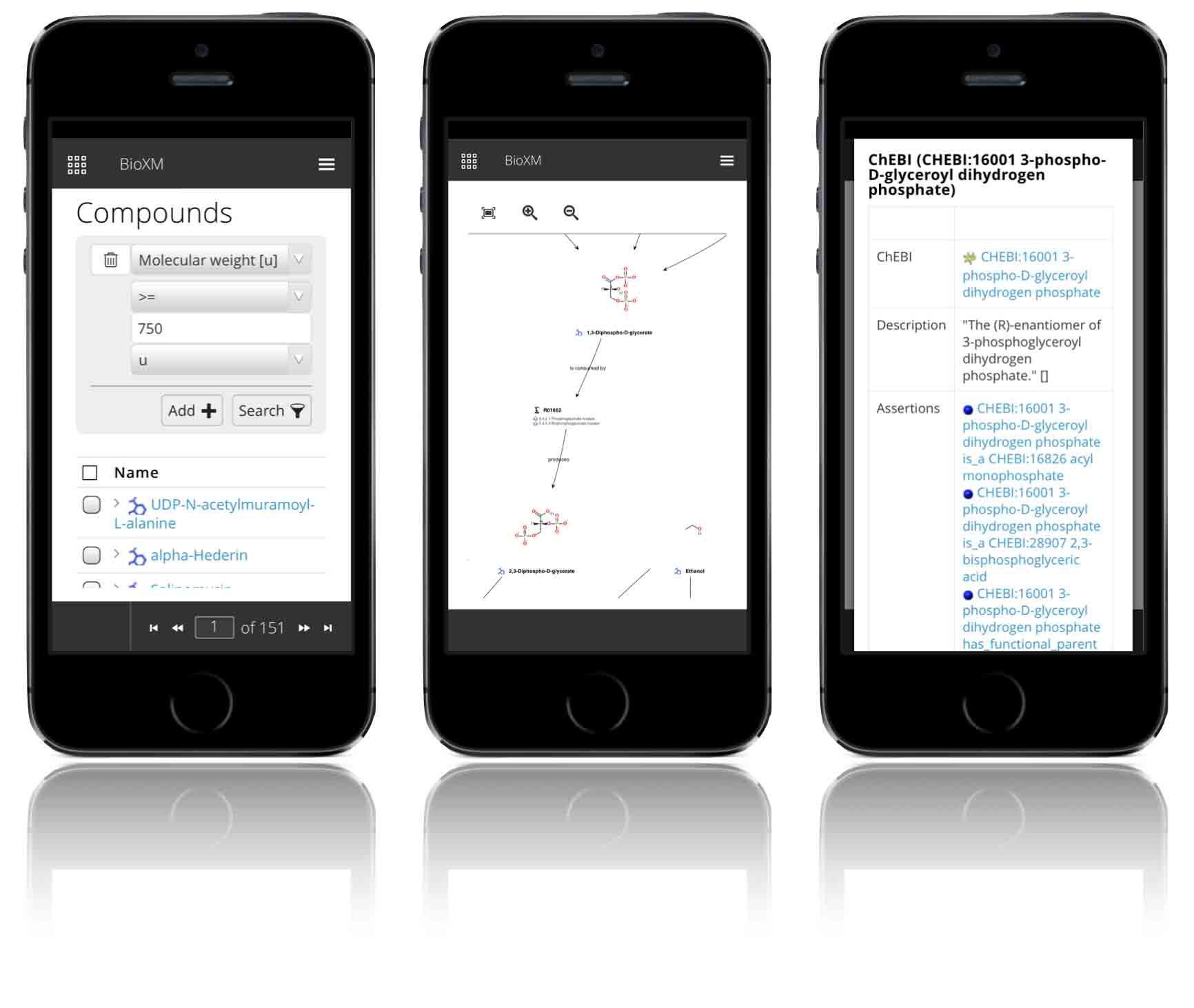
The BioXM system can be used to create browser-based web apps that dynamically access the semantic knowledge network on any device.
In this way, easy-to-use frontends can be created to ensure that any kind of user — from novice to expert — can contribute to and benefit from the organization’s collective knowledge.
References
Back to top